33. Covariate indipendenti dal tempo#
Partendo dai modelli di crescita lineare descritti nei capitoli precedenti, ci chiediamo se le differenze individuali nelle traiettorie di cambiamento siano associate ad altre variabili. Questo capitolo introduce modelli che esaminano se la variabilità nell’intercetta e nella pendenza del modello di crescita lineare può essere spiegata da covariate invarianti nel tempo. Queste sono variabili a livello di persona che non cambiano nel tempo, come il genere, le condizioni sperimentali e lo stato socio-economico. Nel contesto dei modelli a crescita latente, tali variabili vengono considerate come variabili indipendenti in un modello di regressione multipla in cui l’intercetta e la pendenza del modello di crescita lineare sono le variabili dipendenti. Le covariate invarianti nel tempo possono essere continue, ordinali o categoriche.
Studiando come le differenze individuali nell’intercetta e nella pendenza sono correlate ad altre variabili a livello della persona, possiamo comprendere le possibili ragioni per cui gli individui cambiano in modi diversi. Tuttavia, i risultati di questi modelli non forniscono ulteriori inferenze sulla causalità rispetto ai tipici modelli di regressione. Gli utenti dovrebbero essere cauti quando discutono tali effetti e tenere presente se la covariata invariante nel tempo è stata assegnata in maniera casuale nel contesto di un protocollo sperimentale, oppure se i dati provengono da ricerche osservazionali.
Per questo esempio considereremo i dati di prestazione matematica dal data set NLSY-CYA Long Data [si veda Grimm et al. [GRE16]]. Iniziamo a leggere i dati.
# set filepath for data file
filepath <- "https://raw.githubusercontent.com/LRI-2/Data/main/GrowthModeling/nlsy_math_long_R.dat"
# read in the text data file using the url() function
dat <- read.table(
file = url(filepath),
na.strings = "."
) # indicates the missing data designator
# copy data with new name
nlsy_math_long <- dat
# Add names the columns of the data set
names(nlsy_math_long) <- c(
"id", "female", "lb_wght",
"anti_k1", "math", "grade",
"occ", "age", "men",
"spring", "anti"
)
# view the first few observations in the data set
head(nlsy_math_long, 10)
| id | female | lb_wght | anti_k1 | math | grade | occ | age | men | spring | anti | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| <int> | <int> | <int> | <int> | <int> | <int> | <int> | <int> | <int> | <int> | <int> | |
| 1 | 201 | 1 | 0 | 0 | 38 | 3 | 2 | 111 | 0 | 1 | 0 |
| 2 | 201 | 1 | 0 | 0 | 55 | 5 | 3 | 135 | 1 | 1 | 0 |
| 3 | 303 | 1 | 0 | 1 | 26 | 2 | 2 | 121 | 0 | 1 | 2 |
| 4 | 303 | 1 | 0 | 1 | 33 | 5 | 3 | 145 | 0 | 1 | 2 |
| 5 | 2702 | 0 | 0 | 0 | 56 | 2 | 2 | 100 | NA | 1 | 0 |
| 6 | 2702 | 0 | 0 | 0 | 58 | 4 | 3 | 125 | NA | 1 | 2 |
| 7 | 2702 | 0 | 0 | 0 | 80 | 8 | 4 | 173 | NA | 1 | 2 |
| 8 | 4303 | 1 | 0 | 0 | 41 | 3 | 2 | 115 | 0 | 0 | 1 |
| 9 | 4303 | 1 | 0 | 0 | 58 | 4 | 3 | 135 | 0 | 1 | 2 |
| 10 | 5002 | 0 | 0 | 4 | 46 | 4 | 2 | 117 | NA | 1 | 4 |
nlsy_math_long |>
ggplot(
aes(grade, math, group = id)
) +
geom_line(alpha = 0.1) + # add individual line with transparency
stat_summary( # add average line
aes(group = 1),
fun = mean,
geom = "line",
linewidth = 1.5,
color = "red"
) +
labs(x = "Grade at testing", y = "PAT Mathematics")
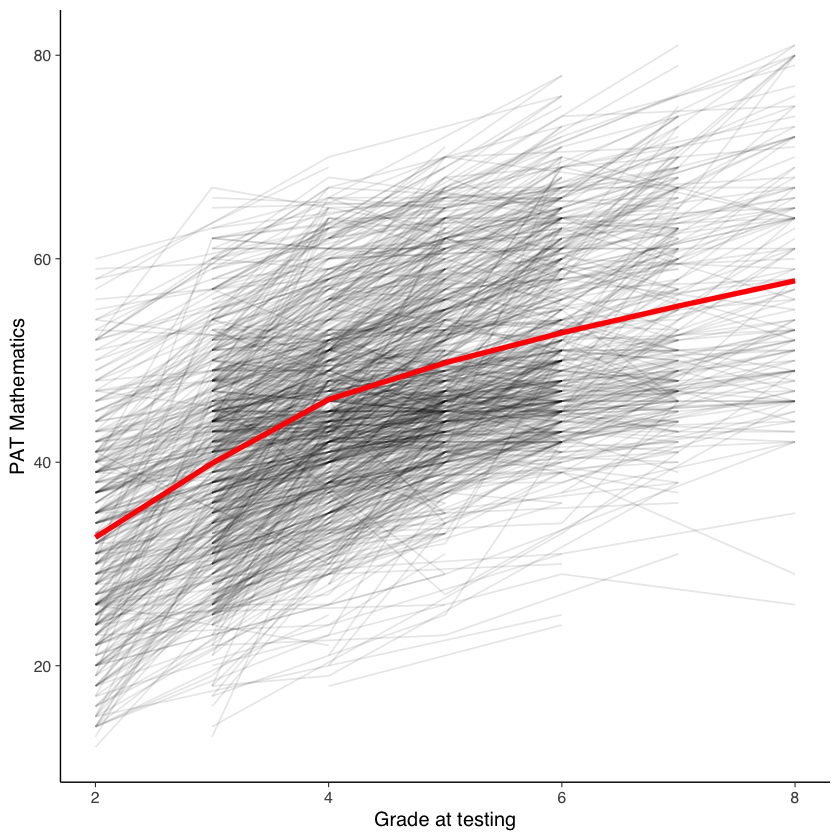
Per vedere il cambiamento a livello individuale, possiamo campionare 20 individui e rappresentare per ognuno il loro cambiamento nel punteggio di matematica.
# sample 20 ids
people <- unique(nlsy_math_long$id) %>% sample(20)
# do separate graph for each individual
nlsy_math_long %>%
filter(id %in% people) %>% # filter only sampled cases
ggplot(aes(grade, math, group = 1)) +
geom_line() +
facet_wrap(~id) + # a graph for each individual
labs(x = "Grade at testing", y = "PAT Mathematics")
`geom_line()`: Each group consists of only one observation.
ℹ Do you need to adjust the group aesthetic?
`geom_line()`: Each group consists of only one observation.
ℹ Do you need to adjust the group aesthetic?
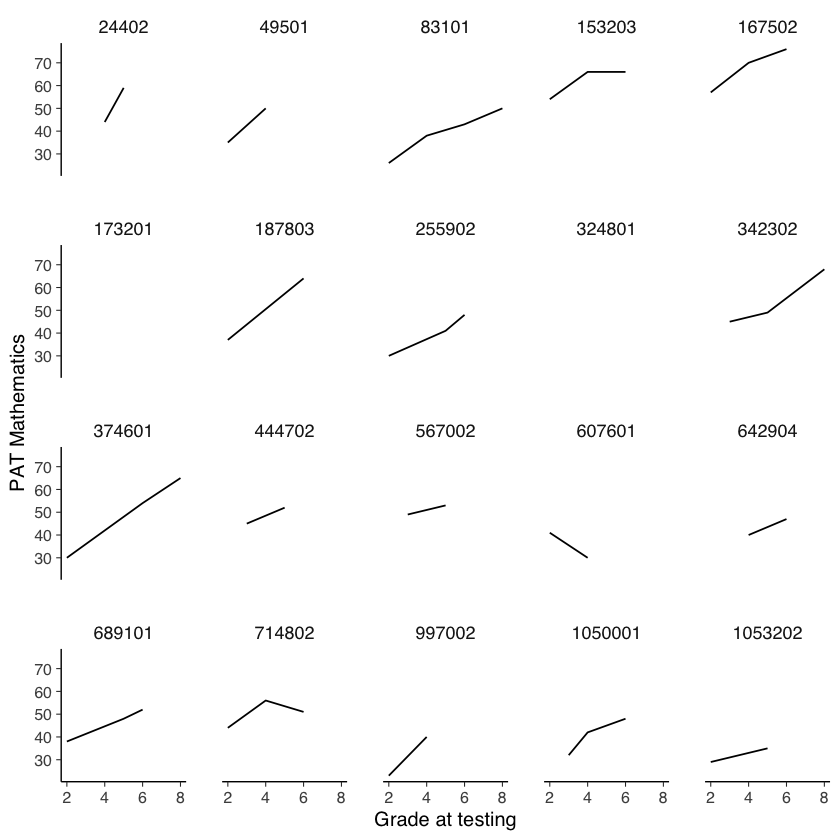
Per semplicità, leggiamo gli stessi dati in formato wide da un file.
# set filepath for data file
filepath <- "https://raw.githubusercontent.com/LRI-2/Data/main/GrowthModeling/nlsy_math_wide_R.dat"
# read in the text data file using the url() function
dat <- read.table(
file = url(filepath),
na.strings = "."
) # indicates the missing data designator
# copy data with new name
nlsy_math_wide <- dat
# Give the variable names
names(nlsy_math_wide) <- c(
"id", "female", "lb_wght", "anti_k1",
"math2", "math3", "math4", "math5", "math6", "math7", "math8",
"age2", "age3", "age4", "age5", "age6", "age7", "age8",
"men2", "men3", "men4", "men5", "men6", "men7", "men8",
"spring2", "spring3", "spring4", "spring5", "spring6", "spring7", "spring8",
"anti2", "anti3", "anti4", "anti5", "anti6", "anti7", "anti8"
)
# view the first few observations (and columns) in the data set
head(nlsy_math_wide[, 1:11], 10)
| id | female | lb_wght | anti_k1 | math2 | math3 | math4 | math5 | math6 | math7 | math8 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| <int> | <int> | <int> | <int> | <int> | <int> | <int> | <int> | <int> | <int> | <int> | |
| 1 | 201 | 1 | 0 | 0 | NA | 38 | NA | 55 | NA | NA | NA |
| 2 | 303 | 1 | 0 | 1 | 26 | NA | NA | 33 | NA | NA | NA |
| 3 | 2702 | 0 | 0 | 0 | 56 | NA | 58 | NA | NA | NA | 80 |
| 4 | 4303 | 1 | 0 | 0 | NA | 41 | 58 | NA | NA | NA | NA |
| 5 | 5002 | 0 | 0 | 4 | NA | NA | 46 | NA | 54 | NA | 66 |
| 6 | 5005 | 1 | 0 | 0 | 35 | NA | 50 | NA | 60 | NA | 59 |
| 7 | 5701 | 0 | 0 | 2 | NA | 62 | 61 | NA | NA | NA | NA |
| 8 | 6102 | 0 | 0 | 0 | NA | NA | 55 | 67 | NA | 81 | NA |
| 9 | 6801 | 1 | 0 | 0 | NA | 54 | NA | 62 | NA | 66 | NA |
| 10 | 6802 | 0 | 0 | 0 | NA | 55 | NA | 66 | NA | 68 | NA |
Specifichiamo il modello SEM (si noti che, anche in questo caso, la scrittura del modello può essere semplificata usando la funzione growth).
#writing out linear growth model with tic in full SEM way
lg_math_tic_lavaan_model <- '
#latent variable definitions
#intercept
eta1 =~ 1*math2+
1*math3+
1*math4+
1*math5+
1*math6+
1*math7+
1*math8
#linear slope
eta2 =~ 0*math2+
1*math3+
2*math4+
3*math5+
4*math6+
5*math7+
6*math8
#factor variances
eta1 ~~ eta1
eta2 ~~ eta2
#factor covariance
eta1 ~~ eta2
#manifest variances (set equal by naming theta)
math2 ~~ theta*math2
math3 ~~ theta*math3
math4 ~~ theta*math4
math5 ~~ theta*math5
math6 ~~ theta*math6
math7 ~~ theta*math7
math8 ~~ theta*math8
#latent means (freely estimated)
eta1 ~ 1
eta2 ~ 1
#manifest means (fixed to zero)
math2 ~ 0*1
math3 ~ 0*1
math4 ~ 0*1
math5 ~ 0*1
math6 ~ 0*1
math7 ~ 0*1
math8 ~ 0*1
#Time invariant covaraite
#regression of time-invariant covariate on intercept and slope factors
eta1 ~ lb_wght + anti_k1
eta2 ~ lb_wght + anti_k1
#variance of TIV covariates
lb_wght ~~ lb_wght
anti_k1 ~~ anti_k1
#covariance of TIV covaraites
lb_wght ~~ anti_k1
#means of TIV covariates (freely estimated)
lb_wght ~ 1
anti_k1 ~ 1
' #end of model definition
Adattiamo il modello ai dati.
lg_math_tic_lavaan_fit <- sem(lg_math_tic_lavaan_model,
data = nlsy_math_wide,
meanstructure = TRUE,
estimator = "ML",
missing = "fiml"
)
Warning message in lav_data_full(data = data, group = group, cluster = cluster, :
“lavaan WARNING: some observed variances are (at least) a factor 1000 times larger than others; use varTable(fit) to investigate”
Warning message in lav_data_full(data = data, group = group, cluster = cluster, :
“lavaan WARNING:
due to missing values, some pairwise combinations have less than
10% coverage; use lavInspect(fit, "coverage") to investigate.”
Warning message in lav_mvnorm_missing_h1_estimate_moments(Y = X[[g]], wt = WT[[g]], :
“lavaan WARNING:
Maximum number of iterations reached when computing the sample
moments using EM; use the em.h1.iter.max= argument to increase the
number of iterations”
Esaminiamo la soluzione ottenuta.
summary(lg_math_tic_lavaan_fit, fit.measures = TRUE) |>
print()
lavaan 0.6.15 ended normally after 107 iterations
Estimator ML
Optimization method NLMINB
Number of model parameters 21
Number of equality constraints 6
Number of observations 933
Number of missing patterns 61
Model Test User Model:
Test statistic 220.221
Degrees of freedom 39
P-value (Chi-square) 0.000
Model Test Baseline Model:
Test statistic 892.616
Degrees of freedom 36
P-value 0.000
User Model versus Baseline Model:
Comparative Fit Index (CFI) 0.788
Tucker-Lewis Index (TLI) 0.805
Robust Comparative Fit Index (CFI) 0.920
Robust Tucker-Lewis Index (TLI) 0.926
Loglikelihood and Information Criteria:
Loglikelihood user model (H0) -9785.085
Loglikelihood unrestricted model (H1) -9674.975
Akaike (AIC) 19600.171
Bayesian (BIC) 19672.747
Sample-size adjusted Bayesian (SABIC) 19625.108
Root Mean Square Error of Approximation:
RMSEA 0.071
90 Percent confidence interval - lower 0.062
90 Percent confidence interval - upper 0.080
P-value H_0: RMSEA <= 0.050 0.000
P-value H_0: RMSEA >= 0.080 0.046
Robust RMSEA 0.100
90 Percent confidence interval - lower 0.000
90 Percent confidence interval - upper 0.183
P-value H_0: Robust RMSEA <= 0.050 0.218
P-value H_0: Robust RMSEA >= 0.080 0.654
Standardized Root Mean Square Residual:
SRMR 0.097
Parameter Estimates:
Standard errors Standard
Information Observed
Observed information based on Hessian
Latent Variables:
Estimate Std.Err z-value P(>|z|)
eta1 =~
math2 1.000
math3 1.000
math4 1.000
math5 1.000
math6 1.000
math7 1.000
math8 1.000
eta2 =~
math2 0.000
math3 1.000
math4 2.000
math5 3.000
math6 4.000
math7 5.000
math8 6.000
Regressions:
Estimate Std.Err z-value P(>|z|)
eta1 ~
lb_wght -2.716 1.294 -2.099 0.036
anti_k1 -0.551 0.232 -2.369 0.018
eta2 ~
lb_wght 0.625 0.333 1.873 0.061
anti_k1 -0.019 0.059 -0.327 0.743
Covariances:
Estimate Std.Err z-value P(>|z|)
.eta1 ~~
.eta2 -0.078 1.145 -0.068 0.945
lb_wght ~~
anti_k1 0.007 0.014 0.548 0.584
Intercepts:
Estimate Std.Err z-value P(>|z|)
.eta1 36.290 0.497 73.052 0.000
.eta2 4.315 0.122 35.420 0.000
.math2 0.000
.math3 0.000
.math4 0.000
.math5 0.000
.math6 0.000
.math7 0.000
.math8 0.000
lb_wght 0.080 0.009 9.031 0.000
anti_k1 1.454 0.050 29.216 0.000
Variances:
Estimate Std.Err z-value P(>|z|)
.eta1 63.064 5.609 11.242 0.000
.eta2 0.713 0.326 2.185 0.029
.math2 (thet) 36.257 1.868 19.406 0.000
.math3 (thet) 36.257 1.868 19.406 0.000
.math4 (thet) 36.257 1.868 19.406 0.000
.math5 (thet) 36.257 1.868 19.406 0.000
.math6 (thet) 36.257 1.868 19.406 0.000
.math7 (thet) 36.257 1.868 19.406 0.000
.math8 (thet) 36.257 1.868 19.406 0.000
lb_wght 0.074 0.003 21.599 0.000
anti_k1 2.312 0.107 21.599 0.000
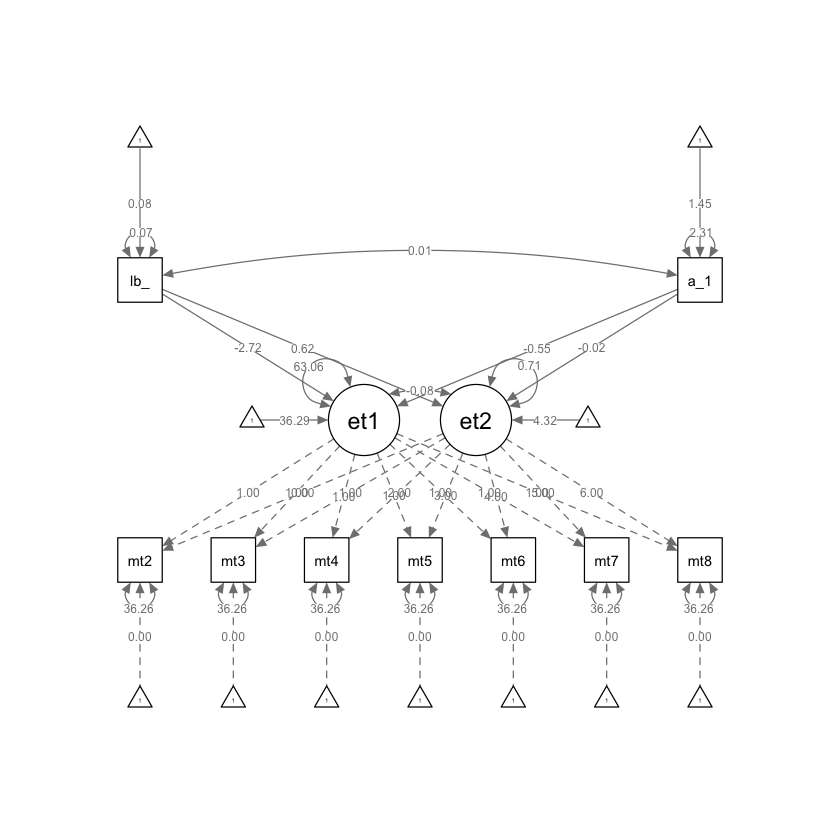
Creiamo un diagramma di percorso.
semPaths(lg_math_tic_lavaan_fit,what = "path", whatLabels = "par")
33.1. Valutare il contributo delle covariate#
Per valutare se le covariate considerate migliorano l’adattamento del modello, confronteremo il modello precedente con un modello vincolato in cui l’effetto delle covariate viene fissato a zero.
#writing out linear growth model with tic in full SEM way
lg_math_ticZERO_lavaan_model <- '
#latent variable definitions
#intercept
eta1 =~ 1*math2+
1*math3+
1*math4+
1*math5+
1*math6+
1*math7+
1*math8
#linear slope
eta2 =~ 0*math2+
1*math3+
2*math4+
3*math5+
4*math6+
5*math7+
6*math8
#factor variances
eta1 ~~ eta1
eta2 ~~ eta2
#factor covariance
eta1 ~~ eta2
#manifest variances (set equal by naming theta)
math2 ~~ theta*math2
math3 ~~ theta*math3
math4 ~~ theta*math4
math5 ~~ theta*math5
math6 ~~ theta*math6
math7 ~~ theta*math7
math8 ~~ theta*math8
#latent means (freely estimated)
eta1 ~ 1
eta2 ~ 1
#manifest means (fixed to zero)
math2 ~ 0*1
math3 ~ 0*1
math4 ~ 0*1
math5 ~ 0*1
math6 ~ 0*1
math7 ~ 0*1
math8 ~ 0*1
#Time invariant covaraite
#regression of time-invariant covariate on intercept and slope factors
#FIXED to 0
eta1 ~ 0*lb_wght + 0*anti_k1
eta2 ~ 0*lb_wght + 0*anti_k1
#variance of TIV covariates
lb_wght ~~ lb_wght
anti_k1 ~~ anti_k1
#covariance of TIV covaraites
lb_wght ~~ anti_k1
#means of TIV covariates (freely estimated)
lb_wght ~ 1
anti_k1 ~ 1
' #end of model definition
Adattiamo il modello ai dati.
lg_math_ticZERO_lavaan_fit <- sem(lg_math_ticZERO_lavaan_model,
data = nlsy_math_wide,
meanstructure = TRUE,
estimator = "ML",
missing = "fiml"
)
Warning message in lav_data_full(data = data, group = group, cluster = cluster, :
“lavaan WARNING: some observed variances are (at least) a factor 1000 times larger than others; use varTable(fit) to investigate”
Warning message in lav_data_full(data = data, group = group, cluster = cluster, :
“lavaan WARNING:
due to missing values, some pairwise combinations have less than
10% coverage; use lavInspect(fit, "coverage") to investigate.”
Warning message in lav_mvnorm_missing_h1_estimate_moments(Y = X[[g]], wt = WT[[g]], :
“lavaan WARNING:
Maximum number of iterations reached when computing the sample
moments using EM; use the em.h1.iter.max= argument to increase the
number of iterations”
Esaminiamo il risultato ottenuto.
summary(lg_math_ticZERO_lavaan_fit, fit.measures = TRUE) |>
print()
lavaan 0.6.15 ended normally after 92 iterations
Estimator ML
Optimization method NLMINB
Number of model parameters 17
Number of equality constraints 6
Number of observations 933
Number of missing patterns 61
Model Test User Model:
Test statistic 234.467
Degrees of freedom 43
P-value (Chi-square) 0.000
Model Test Baseline Model:
Test statistic 892.616
Degrees of freedom 36
P-value 0.000
User Model versus Baseline Model:
Comparative Fit Index (CFI) 0.776
Tucker-Lewis Index (TLI) 0.813
Robust Comparative Fit Index (CFI) 0.917
Robust Tucker-Lewis Index (TLI) 0.931
Loglikelihood and Information Criteria:
Loglikelihood user model (H0) -9792.208
Loglikelihood unrestricted model (H1) -9674.975
Akaike (AIC) 19606.416
Bayesian (BIC) 19659.638
Sample-size adjusted Bayesian (SABIC) 19624.703
Root Mean Square Error of Approximation:
RMSEA 0.069
90 Percent confidence interval - lower 0.061
90 Percent confidence interval - upper 0.078
P-value H_0: RMSEA <= 0.050 0.000
P-value H_0: RMSEA >= 0.080 0.020
Robust RMSEA 0.097
90 Percent confidence interval - lower 0.000
90 Percent confidence interval - upper 0.173
P-value H_0: Robust RMSEA <= 0.050 0.208
P-value H_0: Robust RMSEA >= 0.080 0.646
Standardized Root Mean Square Residual:
SRMR 0.103
Parameter Estimates:
Standard errors Standard
Information Observed
Observed information based on Hessian
Latent Variables:
Estimate Std.Err z-value P(>|z|)
eta1 =~
math2 1.000
math3 1.000
math4 1.000
math5 1.000
math6 1.000
math7 1.000
math8 1.000
eta2 =~
math2 0.000
math3 1.000
math4 2.000
math5 3.000
math6 4.000
math7 5.000
math8 6.000
Regressions:
Estimate Std.Err z-value P(>|z|)
eta1 ~
lb_wght 0.000
anti_k1 0.000
eta2 ~
lb_wght 0.000
anti_k1 0.000
Covariances:
Estimate Std.Err z-value P(>|z|)
.eta1 ~~
.eta2 -0.181 1.150 -0.158 0.875
lb_wght ~~
anti_k1 0.007 0.014 0.548 0.584
Intercepts:
Estimate Std.Err z-value P(>|z|)
.eta1 35.267 0.355 99.229 0.000
.eta2 4.339 0.088 49.136 0.000
.math2 0.000
.math3 0.000
.math4 0.000
.math5 0.000
.math6 0.000
.math7 0.000
.math8 0.000
lb_wght 0.080 0.009 9.031 0.000
anti_k1 1.454 0.050 29.216 0.000
Variances:
Estimate Std.Err z-value P(>|z|)
.eta1 64.562 5.659 11.408 0.000
.eta2 0.733 0.327 2.238 0.025
.math2 (thet) 36.230 1.867 19.410 0.000
.math3 (thet) 36.230 1.867 19.410 0.000
.math4 (thet) 36.230 1.867 19.410 0.000
.math5 (thet) 36.230 1.867 19.410 0.000
.math6 (thet) 36.230 1.867 19.410 0.000
.math7 (thet) 36.230 1.867 19.410 0.000
.math8 (thet) 36.230 1.867 19.410 0.000
lb_wght 0.074 0.003 21.599 0.000
anti_k1 2.312 0.107 21.599 0.000
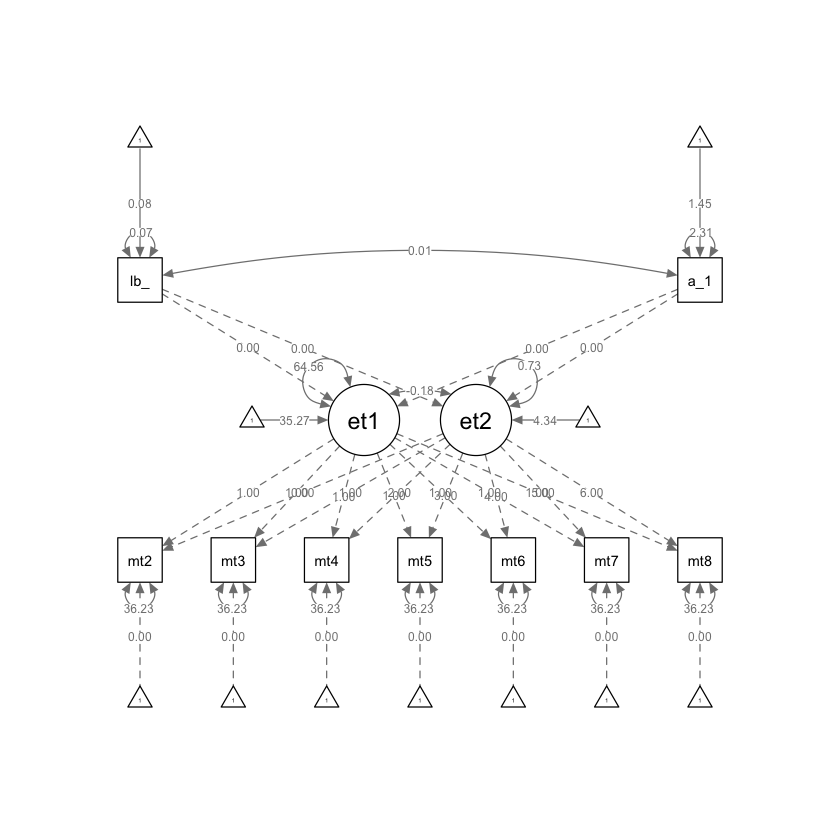
Generiamo il diagramma di percorso.
semPaths(lg_math_ticZERO_lavaan_fit, what = "path", whatLabels = "par")
Eseguiamo il confronto tra i due modelli mediante il test del rapporto tra verosimiglianze.
lavTestLRT(lg_math_tic_lavaan_fit, lg_math_ticZERO_lavaan_fit) |>
print()
Chi-Squared Difference Test
Df AIC BIC Chisq Chisq diff RMSEA Df diff
lg_math_tic_lavaan_fit 39 19600 19673 220.22
lg_math_ticZERO_lavaan_fit 43 19606 19660 234.47 14.245 0.052395 4
Pr(>Chisq)
lg_math_tic_lavaan_fit
lg_math_ticZERO_lavaan_fit 0.006552 **
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Il valore-p associato alla statistica chi-quadrato relativa alla differenza tra i due modelli è piccolo. Questo significa che i vincoli aggiuntivi hanno impattato in maniera sostanziale la bontà di adattamento del modello ai dati. Da ciò concludiamo che le covariate forniscono un contributo utile al modello.
33.2. Confronto con il modello misto#
Eseguiamo l’analisi utilizzando un modello misto con intercetta e pendenza casuale. Confronteremo un modello ridotto, che include solo l’effetto del tempo, con un modello completo che include le covariate esaminate in precedenza. Il modello completo include gli effetti principali delle covariate e l’interazione tra le covariate e il tempo.
Adattiamo il modello “completo”.
nlsy_math_long$grade_c2 <- nlsy_math_long$grade-2
fit2_lmer <- lmer(
math ~ 1 + grade_c2 + lb_wght + anti_k1 + I(grade_c2 * lb_wght) + I(grade_c2 * anti_k1) +
(1 + grade_c2 | id),
data = nlsy_math_long,
REML = FALSE,
na.action = na.exclude
)
Warning message in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
“Model failed to converge with max|grad| = 0.00577356 (tol = 0.002, component 1)”
Esaminiamo i risultati ottenuti.
summary(fit2_lmer)
Linear mixed model fit by maximum likelihood ['lmerMod']
Formula: math ~ 1 + grade_c2 + lb_wght + anti_k1 + I(grade_c2 * lb_wght) +
I(grade_c2 * anti_k1) + (1 + grade_c2 | id)
Data: nlsy_math_long
AIC BIC logLik deviance df.resid
15943.1 16000.2 -7961.6 15923.1 2211
Scaled residuals:
Min 1Q Median 3Q Max
-3.07986 -0.52517 -0.00867 0.53079 2.53455
Random effects:
Groups Name Variance Std.Dev. Corr
id (Intercept) 63.0652 7.941
grade_c2 0.7141 0.845 -0.01
Residual 36.2543 6.021
Number of obs: 2221, groups: id, 932
Fixed effects:
Estimate Std. Error t value
(Intercept) 36.28983 0.49630 73.120
grade_c2 4.31521 0.12060 35.782
lb_wght -2.71621 1.29359 -2.100
anti_k1 -0.55087 0.23246 -2.370
I(grade_c2 * lb_wght) 0.62463 0.33314 1.875
I(grade_c2 * anti_k1) -0.01930 0.05886 -0.328
Correlation of Fixed Effects:
(Intr) grd_c2 lb_wgh ant_k1 I(_2*l
grade_c2 -0.529
lb_wght -0.194 0.096
anti_k1 -0.671 0.358 -0.025
I(grd_2*l_) 0.091 -0.168 -0.532 0.026
I(gr_2*a_1) 0.343 -0.660 0.026 -0.529 -0.055
optimizer (nloptwrap) convergence code: 0 (OK)
Model failed to converge with max|grad| = 0.00577356 (tol = 0.002, component 1)
Adattiamo un modello misto vincolato senza covariate, utilizzando un modello con intercetta e pendenza casuale.
fit3_lmer <- lmer(
math ~ 1 + grade_c2 + (1 + grade_c2 | id),
data = nlsy_math_long,
REML = FALSE,
na.action = na.exclude
)
Confrontiamo i due modelli utilizzando il test del rapporto di verosimiglianza.
lavTestLRT(fit2_lmer, fit3_lmer) |>
print()
Error in h(simpleError(msg, call)): error in evaluating the argument 'x' in selecting a method for function 'print': no slot of name "Options" for this object of class "lmerMod"
Traceback:
1. print(lavTestLRT(fit2_lmer, fit3_lmer))
2. lavTestLRT(fit2_lmer, fit3_lmer)
3. .handleSimpleError(function (cond)
. .Internal(C_tryCatchHelper(addr, 1L, cond)), "no slot of name \"Options\" for this object of class \"lmerMod\"",
. base::quote(lavTestLRT(fit2_lmer, fit3_lmer)))
4. h(simpleError(msg, call))
Il risultato è sostanzialmente identico a quello ottenuto con il modello LGM.
